Task
使用朴素贝叶斯模型和高斯判别分析模型对iris分类
导入模块
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import accuracy_score
plt.rcParams['font.sans-serif'] = ['Microsoft YaHei']
%matplotlib inline查看数据
# 读取数据
feat_names = ['sepal-length', 'sepal-width', 'petal-length', 'petal-width', 'species']
dpath = '../data/'
df = pd.read_csv(dpath + 'iris.csv', names=feat_names)
df.head()| sepal-length | sepal-width | petal-length | petal-width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
# 查看数据总体情况
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 sepal-length 150 non-null float64
1 sepal-width 150 non-null float64
2 petal-length 150 non-null float64
3 petal-width 150 non-null float64
4 species 150 non-null object
dtypes: float64(4), object(1)
memory usage: 6.0+ KB
# 查看缺失值情况
column_null = df.isnull().sum(axis=0)
row_null = df.isnull().sum(axis=1)
all_null = df.isnull().sum().sum()
all_null0
# 查看数值型特征的统计量
df.describe()| sepal-length | sepal-width | petal-length | petal-width | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| std | 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| min | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
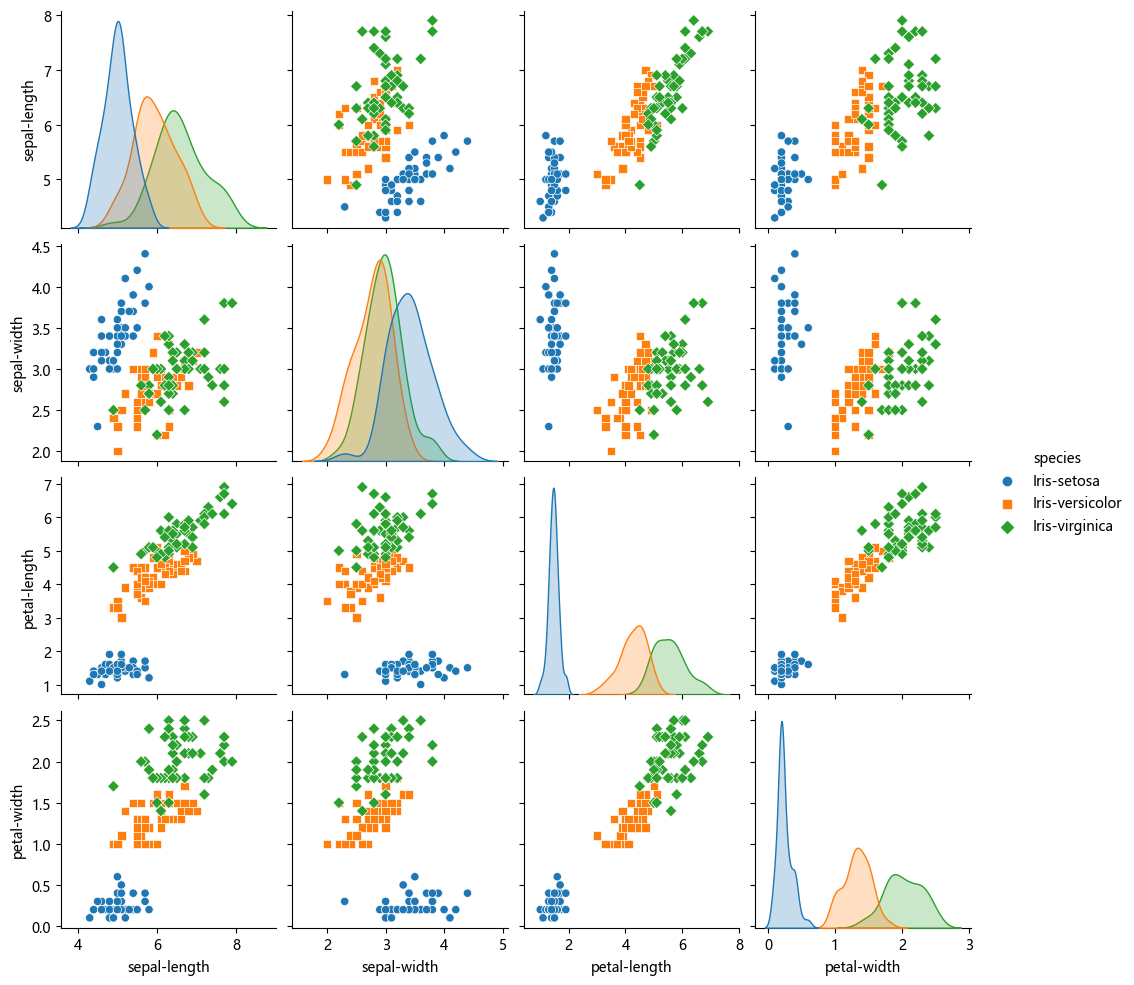
# 查看特征两两之间的散点图
sns.pairplot(df, hue='species', kind='scatter', diag_kind='kde', markers=["o", "s", "D"], diag_kws=dict(fill=True))d:\Anaconda3\Lib\site-packages\seaborn\axisgrid.py:118: UserWarning: The figure layout has changed to tight
self._figure.tight_layout(*args, **kwargs)
<seaborn.axisgrid.PairGrid at 0x2f6a5635e10>
数据预处理
# 将标签字符串映射为整数
target_map = {'Iris-setosa':0,
'Iris-versicolor':1,
'Iris-virginica':2 }
df['species'] = df['species'].apply(lambda x: target_map[x])
df.head()| sepal-length | sepal-width | petal-length | petal-width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0 |
# 从原始数据分离x, y
y = df['species']
X = df.drop('species', axis=1)
X, y( sepal-length sepal-width petal-length petal-width
0 5.1 3.5 1.4 0.2
1 4.9 3.0 1.4 0.2
2 4.7 3.2 1.3 0.2
3 4.6 3.1 1.5 0.2
4 5.0 3.6 1.4 0.2
.. ... ... ... ...
145 6.7 3.0 5.2 2.3
146 6.3 2.5 5.0 1.9
147 6.5 3.0 5.2 2.0
148 6.2 3.4 5.4 2.3
149 5.9 3.0 5.1 1.8
[150 rows x 4 columns],
0 0
1 0
2 0
3 0
4 0
..
145 2
146 2
147 2
148 2
149 2
Name: species, Length: 150, dtype: int64)
# 数据划分
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)模型训练
朴素贝叶斯模型
朴素贝叶斯模型: https://scikit-learn.org/stable/modules/naive_bayes.html#gaussian-naive-bayes
贝叶斯公式: $P(y|x) = \frac{P(xy)}{P(x)} = \frac{P(x|y)P(y)}{P(x)}$
朴素贝叶斯模型就是根据贝叶斯公式进行设计,inference时对于给定的输入x,将x作为条件,判断每一个标签y的概率。这是一种非常简单而且自然的想法,我们拿到一堆数据去预测另一个未知输入时,不就是在已有的数据里寻找输入和输出之间的潜在规律,然后按照这个规律去看新的输入最大可能的输出是多少。所以训练的目标很简单,就是让预测的概率尽可能地正确。
首先看分母,对于一个输入x,$P(x)$是一个定值,可以不用管。我们只需要把$P(y)$和$P(x|y)$预测好就行了。$P(y)$称为类先验分布,通俗地说是每个类出现的几率。$P(X|y)$称为类条件概率,即在确定类别的条件下,出现这个x的概率。为了估算这两个值,我们需要假设y, x|y服从的分布情况。一般常用的分布有高斯分布、Multinoulli分布等。
值得注意的是,朴素贝叶斯模型假设各个特征之间是相互独立的,这个假设大大简化了模型。尽管这个假设非常强,但在现实世界中朴素贝叶斯模型在很多情况下还是有不错的效果。
from sklearn.naive_bayes import GaussianNB
gnb = GaussianNB()
gnb.fit(X_train, y_train)
y_train_pred = gnb.predict(X_train)
y_test_pred = gnb.predict(X_test)
print('train loss: {}'.format(accuracy_score(y_train_pred, y_train)))
print('test loss: {}'.format(accuracy_score(y_test_pred, y_test)))train loss: 0.9583333333333334
test loss: 0.9666666666666667
print('预测类别: ',gnb.classes_)
print('类先验概率: ',gnb.class_prior_)
print('均值: ', gnb.theta_)
print('方差: ', gnb.var_)预测类别: [0 1 2]
类先验概率: [0.33333333 0.33333333 0.33333333]
均值: [[4.985 3.4025 1.48 0.2525]
[5.93 2.75 4.2525 1.32 ]
[6.61 2.98 5.58 2.04 ]]
方差: [[0.092775 0.15674375 0.0251 0.01349375]
[0.2216 0.093 0.19149375 0.0341 ]
[0.4574 0.1221 0.3236 0.0704 ]]
二次判别分析 Quadratic Discriminant Analysis
二次判别分析:https://scikit-learn.org/stable/modules/lda_qda.html
朴素贝叶斯模型认为各个特征相互独立,二次判别分析则使用多元高斯分布去刻画这些特征分布。
也就是说朴素贝叶斯模型是二次判别分析一种特殊情况。
朴素贝叶斯模型 = QDA + 特征之间独立
LDA = QDA + QDA + 不同类别特征的协方差相同
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
qda.fit(X_train, y_train)
y_train_pred = qda.predict(X_train)
y_test_pred = qda.predict(X_test)
print('train loss: {}'.format(accuracy_score(y_train_pred, y_train)))
print('test loss: {}'.format(accuracy_score(y_test_pred, y_test)))train loss: 0.975
test loss: 1.0
print('类先验:')
print(qda.priors_ )
print('均值:')
print(qda.means_)
print('协方差矩阵:')
print(qda.covariance_)
print('主轴方向:')
print(qda.rotations_)
print("主轴方向的方差:")
print(qda.scalings_)类先验:
[0.33333333 0.33333333 0.33333333]
均值:
[[4.985 3.4025 1.48 0.2525]
[5.93 2.75 4.2525 1.32 ]
[6.61 2.98 5.58 2.04 ]]
协方差矩阵:
[array([[0.09515385, 0.09644872, 0.0094359 , 0.01260256],
[0.09644872, 0.16076282, 0.01569231, 0.01422436],
[0.0094359 , 0.01569231, 0.02574359, 0.00569231],
[0.01260256, 0.01422436, 0.00569231, 0.01383974]]), array([[0.22728205, 0.06538462, 0.14992308, 0.04092308],
[0.06538462, 0.09538462, 0.06269231, 0.03358974],
[0.14992308, 0.06269231, 0.19640385, 0.06046154],
[0.04092308, 0.03358974, 0.06046154, 0.03497436]]), array([[0.46912821, 0.11302564, 0.35123077, 0.04882051],
[0.11302564, 0.12523077, 0.09394872, 0.05235897],
[0.35123077, 0.09394872, 0.33189744, 0.05876923],
[0.04882051, 0.05235897, 0.05876923, 0.07220513]])]
主轴方向:
[array([[ 0.577788 , 0.73618305, -0.31761196, -0.1527029 ],
[ 0.80647248, -0.47185993, 0.35401286, 0.04031434],
[ 0.08971131, -0.48493266, -0.81656606, -0.3000201 ],
[ 0.08783536, -0.01493412, -0.32713515, 0.94076805]]), array([[-0.68454964, 0.60008382, -0.39870293, -0.11102777],
[-0.29380636, -0.70158636, -0.61189706, 0.21687874],
[-0.63479501, -0.22071006, 0.66978334, 0.31577309],
[-0.20519479, -0.31458393, 0.13419481, -0.91701897]]), array([[ 0.74623282, -0.2620598 , 0.43418988, 0.43120806],
[ 0.21974119, 0.7874671 , 0.45198579, -0.35681678],
[ 0.61764038, -0.06722203, -0.59012686, -0.51551125],
[ 0.11563196, 0.5538063 , -0.50885979, 0.64883707]])]
主轴方向的方差:
[array([0.23315722, 0.0268634 , 0.02489103, 0.01058835]), array([0.40663798, 0.07424305, 0.06199862, 0.01116522]), array([0.80068144, 0.11642003, 0.05219651, 0.02916357])]
# 带缩放的QDA
# Regularizes the per-class covariance estimates by transforming S2 as S2 = (1 - reg_param) * S2 + reg_param * np.eye(n_features)
qda = QuadraticDiscriminantAnalysis(reg_param=0.1, store_covariance=True)
qda.fit(X_train, y_train)
y_train_pred = qda.predict(X_train)
y_test_pred = qda.predict(X_test)
print('train loss: {}'.format(accuracy_score(y_train_pred, y_train)))
print('test loss: {}'.format(accuracy_score(y_test_pred, y_test)))train loss: 0.975
test loss: 0.9666666666666667

